For performing quantitative microbiological risk assessment (QMRA) the open-source R package kwb.qmra (available at: Github and Zenodo) was developed at KWB within the DEMOWARE project.
With the kwb.qmra package a QMRA was performed for the Old Ford site within the DEMOWARE project in order to assesss different wastewater reuse options (for details see: Zenodo).
The general workflow for using the R package kwb.qmra is shown in Fig. 1 below.
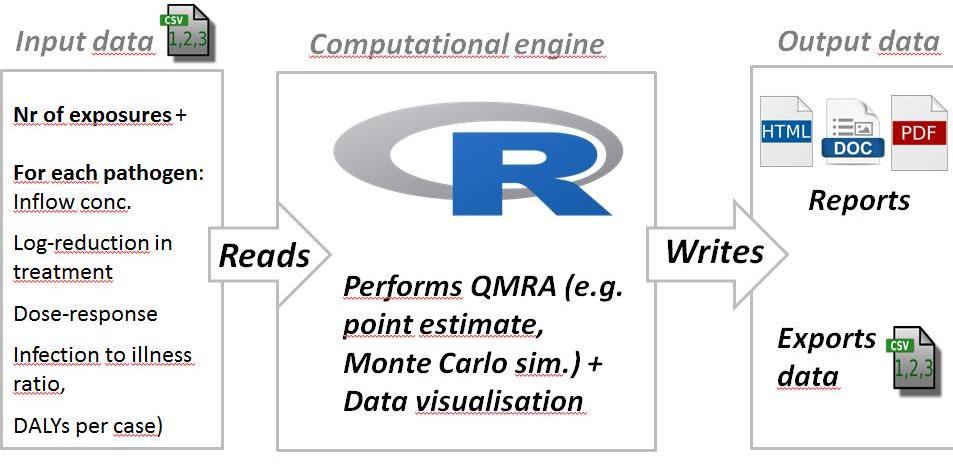
Fig 1: General workflow
The QMRA calculation is based on input data which are stored in six different .CSV configuration files as shown in Fig. 2 below.
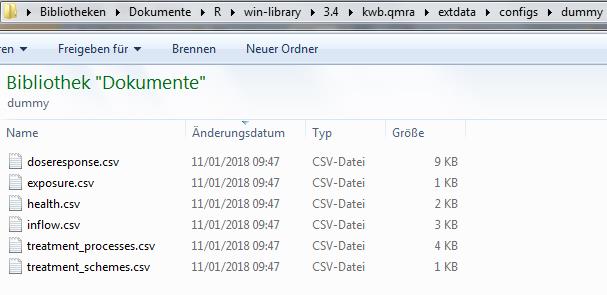
Fig. 2: Screenshot of the six required .CSV configuration files
These contain the required information for:
Inflow concentrations (defined in
inflow.csv): inflow concentrations (fixed value or following a predefined distribution: e.g. min/max values in case of uniform distribution). Inflow concentrations need to be defined for all pathogens that should be used for QMRA.Treatment processes (defined in
treatment_processes.csv): for each treatment process the log-removals for the three different pathogen groups (i.e. Bacteria, Protozoa, Viruses) need to be defined. Again these can be either defined as fixed value or following a used defined distribution (e.g. uniform, which requires specifying min/max values). Ranges for log-removals can be found for example in WHO (2011)Treatment schemes (defined in
treatment_schemes.csv): different treatment processes (defined intreatment_processes.csv) can be combined to treatment schemes (e.g. bankfiltration + slow sand filtration). All defined treatment schemes will be considered in the QMRA calculation.Exposure scenario (defined in
exposure.csv): i.e. the number of exposures per year and the ingested volume per event, which can be defined as fixed value or by following a predefined distribution (e.g. uniform, triangle). In case of a
Monte-Carlo simulation the number of random distribution generations can be specified here also.Dose-response modelling (defined in
doseresponse.csv): the best-fit model (i.e.
exponential or beta-poisson) and its parameters for each pathogen are taken from QMRAwiki (2016).Health parameters (defined in
health.csv): for all pathogens to be used for QMRA the infection to illness factor and the disability-adjusted life years (DALY) per case need to be defined. Values for these parameters can be found for example in WHO (2011)