Workflow: Update Database
Michael Rustler
2021-02-19
Source:vignettes/update_database.Rmd
update_database.RmdInstallation
###############################################################################
### 1 Install R Package Downloading and Installing from GitHub
###############################################################################
install.packages("remotes", repos = "https://cloud.r-project.org")
#Sys.setenv(GITHUB_PAT = "mysecret_access_token")
remotes::install_github("r-lib/remotes@18c7302637053faf21c5b025e1e9243962db1bdc")
###############################################################################
### 2 Install R Package qmra.db (with ACCESS DB)
###############################################################################
### 2.1 Specific Release ACCESS DB from our GitHub account, e.g.
remotes::install_github("kwb-r/qmra.db@0.9.0")
### 2.2 the Latest Development Version
#remotes::install_github("kwb-r/qmra.db") Update Database
Where to find it ?
Run the following command to find out the path to the MS ACCESS Database:
mdb_path <- normalizePath(system.file("database/qmra-db.accdb",
package = "qmra.db"))
mdb_pathHave a look at the database structure
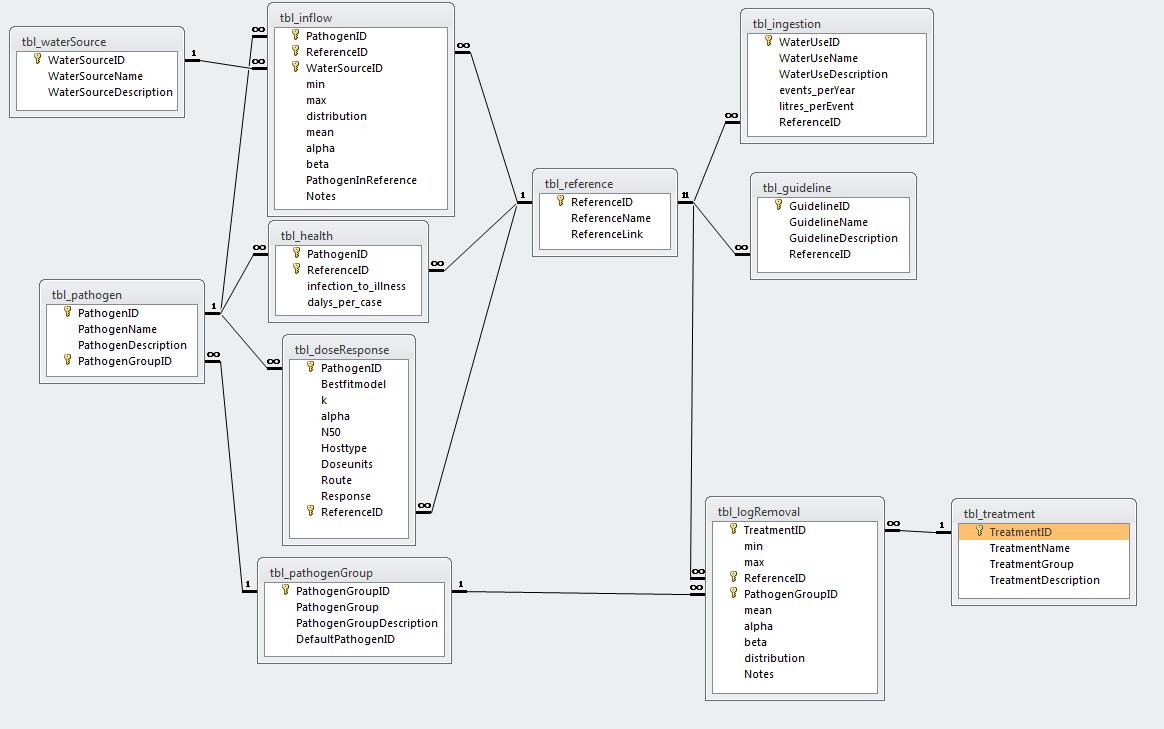
Fig. 1: Screenshot of MS ACCESS Database Structure (might be out of date!)
Make changes
For making changes in the MS ACCESS database the following workflow should be followed:
Open
C:\Users\<yourusername>\Documents\R\win-library\3.5\qmra.db\database\qmra-db.accbwith MS ACCESS,Add your changes and finally
Save it under the same path
C:\Users\<yourusername>\Documents\R\win-library\3.5\qmra.db\database\qmra-db.accb(by overwriting the old one!)
Dump Content in CSV files
Finally please run following R code in order to export the whole content:
qmra.db::dump_access_db(mdb_path)Exporting MS Access Database content into .csv files ...
Running SQL: SELECT * FROM tbl_doseResponse WHERE TRUE
The query returned 37 records with 10 fields: PathogenID,Bestfitmodel,k,alpha,N50,Hosttype,Doseunits,Route,Response,ReferenceID
Running SQL: SELECT * FROM tbl_guideline WHERE TRUE
The query returned 2 records with 4 fields: GuidelineID,GuidelineName,GuidelineDescription,ReferenceID
Running SQL: SELECT * FROM tbl_health WHERE TRUE
The query returned 3 records with 4 fields: PathogenID,ReferenceID,infection_to_illness,dalys_per_case
Running SQL: SELECT * FROM tbl_inflow WHERE TRUE
The query returned 28 records with 11 fields: PathogenID,ReferenceID,WaterSourceID,min,max,distribution,mean,alpha,beta,PathogenInReference,Notes
Running SQL: SELECT * FROM tbl_ingestion WHERE TRUE
The query returned 8 records with 6 fields: WaterUseID,WaterUseName,WaterUseDescription,events_perYear,litres_perEvent,ReferenceID
Running SQL: SELECT * FROM tbl_logRemoval WHERE TRUE
The query returned 74 records with 10 fields: TreatmentID,min,max,ReferenceID,PathogenGroupID,mean,alpha,beta,distribution,Notes
Running SQL: SELECT * FROM tbl_pathogen WHERE TRUE
The query returned 37 records with 4 fields: PathogenID,PathogenName,PathogenDescription,PathogenGroupID
Running SQL: SELECT * FROM tbl_pathogenGroup WHERE TRUE
The query returned 3 records with 4 fields: PathogenGroupID,PathogenGroup,PathogenGroupDescription,DefaultPathogenID
Running SQL: SELECT * FROM tbl_reference WHERE TRUE
The query returned 50 records with 3 fields: ReferenceID,ReferenceName,ReferenceLink
Running SQL: SELECT * FROM tbl_treatment WHERE TRUE
The query returned 28 records with 4 fields: TreatmentID,TreatmentName,TreatmentGroup,TreatmentDescription
Running SQL: SELECT * FROM tbl_waterSource WHERE TRUE
The query returned 8 records with 3 fields: WaterSourceID,WaterSourceName,WaterSourceDescription
Running SQL: SELECT * FROM qry_treatmentRemovals WHERE TRUE
The query returned 74 records with 7 fields: TreatmentGroup,TreatmentName,PathogenGroup,Min,Max,ReferenceName,ReferenceLink
ok. (7.26s) Create ZIP File
Create a zip file qmra-db.zip with the following content:
Modified MS ACCESS DB (qmra-db.accdb) and
Csv files with the whole content of this database (automatically stored in subdirectory qmra-db_accdb/)
zip_path <- qmra.db::create_zipfile(mdb_path)
cat(zip_path)Suggest Your Changes on GitHub
In case that changes are suggested for the MS ACCESS database the following workflow is proposed:
Create a new issue on GitHub at https://github.com/KWB-R/qmra.db/issues with a title starting with Database Update:
Upload the
qmra-db.zipfile (by dragging into the Comment field) andProvide more detailed explainations in the Comment field
Thank you very much for your suggestions! These will be reviewed by our QMRA experts:
Patrick Smeets from KWR Watercycle Research Institute (KWR): @PatrickSmeetsKWR
Wolfgang Seis from Kompetenzzentrum Wasser Berlin gGmbH (KWB): @wseis